Towards 100,000 CPU Cycle-Scavenging by Genetic Algorithms
Al Globus
CSC at NASA Ames Research Center
September 2001
Abstract
Cycle scavenging systems offer 100s to 100,000s of otherwise-idle CPUs
for embarrassingly parallel computations such as genetic algorithms. While
genetic algorithms are generally easy to parallelize, cycle scavenged resources
come and go at random so some sophistication in necessary, particularly
when hundreds of thousands of CPUs are available.
In this paper we propose a master-slave architecture for multi-objective
genetic algorithms on cycle scavengers. The architecture consists of
slave computations running on computational nodes with relatively small
populations, and a central master managing a large pareto front in a disc-based
relational data base . Each slave runs an individual genetic algorithm
and different slaves use different techniques, evolution-parameters
and/or a subset of the objective functions as determined by the master.
The slaves accept immigrants from the master and, after evolution, the
best individuals emigrate back to the master. Slaves may then get
new immigrants and/or evolution policy to be applied to the existing population.
Allowing slaves to use different evolution techniques and parameters can,
when many CPUs are available, avoid committing to a single evolution concept
for a given problem. A sophisticated master can treat the running
slaves as a population of evolutionary techniques and parameters that can
be evolved.
We examine a web-centric design using standard tools such as web servers,
web browsers, PHP, and mySQL. We also consider the applicability of Information
Power Grid tools such as the Globus (no relation to the author) Toolkit.
We intend to implement this architecture with JavaGenes running on at least
two cycle-scavengers: Condor and United Devices. JavaGenes, a genetic algorithm
code written in Java, will be used to evolve multi-species reactive molecular
force field parameters.
Introduction
NASA has applied genetic algorithms (GAs) to a number of problems.
Examples include, but are not limited to, rocket engine design [Akira and
Meng-Sing 2001], wing design [Holst and Pulliam 2001], planet discovery
[Laughlin and Chambers 2001], circuit design [Lohn and Colombano 1998],
molecular design [Globus, et. al. 1999] and molecular force field parameters
discovery [Globus, et. al. 2001]. GAs are stochastic, embarrassingly parallel,
and can tolerate failures. Thus, GAs are well suited to cycle scavenging
computational environments where computers purchased for interactive use
also run batch jobs nights, weekends, and at other idle times.
The best-known cycle scavenging computation is seti@home [http://setiathome.ssl.berkeley.edu/],
currently the largest computation on the planet. Seti@home was using >3
million computers to achieve 23.37 sustained teraflops/sec (979 lifetime
teraflops) as of September 2001 [http://setiathome.ssl.berkeley.edu/totals.html].
The NASA Advanced Supercomputing Division (NAS [http:www.nas.nasa.gov])
has run genetic algorithms using the Condor [http://www.cs.wisc.edu/condor]
cycle scavenger running on ~350 Sun and SGI workstations. In addition,
NAS indends to us the United Devices [http://www.uniteddevices.com/home.htm]
to run genetic algorithms and other codes on the United Devices MetaProcessor,
which cycle scavenges volunteer PCs connected to the Internet. As of September
2001, the MetaProcessor ran on about 900,000 machines [http://members.ud.com/stats/].
Entropia [http://www.entropia.com] and Parabon [http://www.parabon.com]
provide services similar to United Devices.
This paper explores some of the issues that must be faced when moving
JavaGenes [Globus, et. al. 1999], a genetic algorithm written in Java,
from a few hundred processors on the NAS Condor pool to tens of thousands
of processors on the United Devices MetaProcessor. There are alternatives
to the UD MetaProcessor, but United Devices is interested in providing
their resources to NASA free of charge.
First, we briefly review the underlying technologies: genetic algorithms
and cycle-scavenging.
Genetic Algorithms
Genetic algorithms seek to mimic natural evolution's ability to produce
highly functional objects. Natural evolution produces organisms. GAs produce
sets of parameters, programs, molecular designs, and many other structures.
Genetic algorithms usually solve problems by:
-
Expressing the solution to a problem in a data structure.
-
Randomly generating a population of individual solutions.
-
Repeatedly selecting parent individuals at random with a bias towards
better individuals and applying transmission operators to produce children.
Transmission operators include:
-
Crossover: each of two parents is divided into two parts and one part
from each parent is combined into a child.
-
Mutation: a single "parent" is randomly modified to generate a child.
-
Continuing until an acceptable solution is found or exhaustion sets
in.
Better individuals are determined by a "fitness function." Single objective
fitness functions produce a number (the "fitness") that can be compared
to determine the better individual. Multi-objective fitness functions produce
multiple fitness values for each individuals, and comparing two individuals
is more complex since each individual may be superior in different objectives.
For example, one wing design may have low drag but low lift as well, while
a second wing may have high drag but have very high lift. One cannot trivially
say which wing is better. For multi-objective GAs, one attempts to find
the "pareto front," a set of individuals where, for each individual in
the front no other individual is superior in every objective.
For most GAs, fitness function evaluation is by far the most time consuming
step, and each fitness evaluation is typically completely independent.
This means that the main loop is embarrassingly parallel (except for insertion
and removal from the population). Also, since GAs are stochastic, a single
run rarely tells you much. Just as biologists must conduct experiments
with large numbers of animals, GA practitioners often run many copies of
the same code with the same inputs (except the random number seed) to get
statistically reliable results. Note that if enough copies are run, it
is acceptable to lose some runs to system crashes, machines being turned
off, or whatever.
Cycle-Scavenging
Cycle-scavenging systems typically use machines purchased for other purposes
to run batch jobs at night, weekends, and other idle times. Cycle-scavenging
systems typically gain and lose machines at unpredictable times as interactive
users start or stop using their machines, new machines are purchased, machines
are removed from the network, etc. Most cycle-scavengers will move jobs
from machine to machine as necessary
Condor
Condor [Litzkow, et al. 1988] utilizes a cluster of UNIX and Windows NT
workstations on a network. The NAS Condor pool consists of approximately
350 SGI and Sun workstations purchased and used for software development,
visualization, email, document preparation, etc. Each workstation runs
a daemon that watches user IO and CPU load. When a workstation has been
idle for 2 hours, a job from the batch queue is assigned to the workstation
and will run until the daemon detects a keystroke, mouse motion, or high
non-Condor CPU usage. At that point, the job will be removed from the workstation
and placed back on the batch queue.

| Figure 1: NAS workstation usage for June 2001. Note the regular patterns
of five red peaks when workstations are in use during normal business hours.
The green represents Condor enabled batch computation. Only the relatively
small amount of blue indicates wasted CPU cycles. The tall blue spikes
are upgrade times when Condor calculations were temporarily suspended. |
Since Condor runs on collections of workstations continuously connected
to a network, IO to and from each job can run in the hundreds of megabytes
or even a few gigabytes for long running jobs. However, communication between
jobs is difficult since jobs are always starting and stopping. Condor has
a mechanism to set up master-slave messaging relationships via PVM [Goux,
et. al 2000], but JavaGenes does not use it. MPI is not used since MPI
1 does not allow processes to be added or subtracted from an application.
United Devices
United Devices is one of several companies exploiting the success of seti@home
by developing systems to cycle-scavenge PCs. Motivated by a project to
screen potential cancer drugs, people around the world have volunteered
approximately 900,000 PCs to the United Devices MetaProcessor. The drug
screening runs in background and limits it's memory, disc and CPU usage.
United Devices has signed a Letter of Intent with the NAS to execute NASA
codes on the MetaProcessor at no cost and a Space Act Agreement is under
negotiation. NASA plans to port JavaGenes and other applications to the
MetaProcessor. For JavaGenes, this will require major changes to take advantage
of the much larger number of processors and accommodate slower network
connections. The MetaProcessor is designed for intermittently connected
56Kbaud modems, although perhaps 80% of the PCs in the MetaProcessor are
continuously connected by broadband (DSL, cable modem, T1 lines, etc.).
JavaGenes: Current Status
The JavaGenes project presently runs a genetic algorithm on the NAS Condor
Pool [Globus, et. al. 2000b]. JavaGenes is currently a single objective
GA written in Java which evolves parameters for molecular force fields,
molecules, and digital circuits. For the purposes of this paper, we will
only consider evolving molecular force field parameters. Molecular force
fields are potential functions that calculate the energy of molecules and/or
molecular complexes. The reactive, multi-species potentials of interest
have ~5-50 parameters for two species; thus, JavaGenes individuals consist
of 5-50 floating point numbers. The fitness is the root-mean-square of
the difference of externally calculated energies for a set of molecules
and energies calculated using the parameters of a given individual. External
energies can be calculated by expensive, high-quality quantum codes.
As the best approach to evolving molecular force field parameters is
unknown, JavaGenes is used to conduct experiments in genetic algorithm
techniques and genetic-algorithm-parameters for the search. Most experiments
involve 30-50 jobs using the same GA-parameters (except the random number
seed), which means 90-150 jobs for an experiment involving three conditions.
Individual jobs run from a few hours to a few weeks. Checkpointing is implemented
by JavaGenes and all IO uses the NFS automounter to place results on a
single workstation. Analysis of results is via perl scripts run under cron
late at night. The scripts generate html files with links to gif files
of two dimensional plots. A typical display consists of one plot for each
parameter being evolved. Each plot is the value of the best individual
(vertical axis) vs generation (horizontal axis). Colors indicate different
jobs. Parts of the output are moved to an externally accessible web server
so that analysis can be location independent.
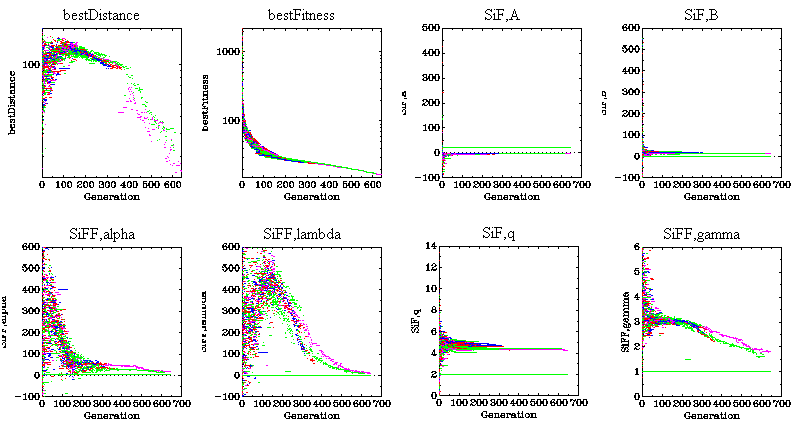
|
FIgure 2: Molecular force field fitness and parameters evolving
over time.
|
Recently, JavaGenes has been modified to pass individuals from one job
into the starting population of another job. This allows a run that
generates, say, Si (silicon) parameters to provide a starting point that
evolves Si and F parameters. However, immigration is accomplished basically
by hand: perl scripts remove the best individual from a job's output files
and format an input file for another job. The second job reads the input
file and modifies the starting population by putting in some number of
immigrants. The second job can alternately use the immigrant to determine,
for all time, some of the parameters. This technique is useful when the
Si parameters have been accurately evolved and can then be used to evolve
Si-F parameters.
New Design
We examine the objectives and criteria for the design. After a brief
discussion of our approach to multi-objective GAs for this project, we
examine a web-centric design based on current standard software and the
status of IPG software with regard to cycle-scavenged computation.
Objectives
The JavaGenes design described below is intended to:
-
Run on both Condor and the UD MetaProcessor with few or no source code
changes and allow for future ports to other platforms (e.g., Entropia and
Parabon).
-
Take advantage of up to 100,000 processors. It is unlikely that the full
MetaProcessor will be available for NASA jobs, so the 100,000 target is
probably more than adequate.
-
Accommodate broadband connected machines only. Machines in the NAS Condor
pool have continuous connectivity to a fast LAN. Approximately 80% of the
UD MetaProcessor compute nodes are broadband connected so it doesn't make
sense to design for 56Kbaud modems..
Criteria
The design should be judged according to the following special criteria
(plus the usual motherhoods):
-
Make appropriate use of cycle-scavenged computational nodes. Since compute
nodes are very plentiful on the MetaProcessor, substantial inefficiencies
may be tolerated to achieve other goals.
-
Tolerate substantial random loss of computational nodes. Compute nodes
on Condor and the MetaProcessor are constantly being turned-off, removed
from the system, etc.
-
Tolerate loss of network connectivity. While we are restricting ourselves
to broadband connectivity, network connectivity can still be lost for a
variety of reasons and this should not be fatal to the computation.
-
Tolerate occasional loss of central facilities. Any central facility will
fail occasionally and this should not fatally affect the computation.
-
Use a multi-objective GA to take advantage of experimental data. The single
objective GA has problems getting the right answers. It is believed that
this can be improved by incorporating experimental data into the fitness
function. It appears that the best way to do this is to use a multiple
objective GA.
-
Require little or no GA parameter/policy tweaking. The bane of GA research
is the large numbers of GA-parameters (e.g., population size, mutation
frequency, etc.) and techniques that must be searched to find the "right"
approach. On the MetaProcessor, we intend to experiment with evolving the
GA-parameter settings while simultaneously searching for the solution.
-
Have security sufficient to run slaves on any machine on the internet.
Anyone on the internet can volunteer their PC to the UD MetaProcessor.
Thus, a variety of security risks must be addressed.
Multi-Objective Approach
JavaGenes molecular force field parameter evolution is expected to have
5-10 objectives. The pareto front in a such a higher dimensional space
is huge and requires many points to sample effectively. However,
the number of immigrants that may be sent from one slave is limited by
bandwidth and master processing power. Therefor, a multi-objective fitness
function for the slaves that identifies one or a few 'best' individuals
is desirable, while a large pareto front of individuals can be managed
by the master on disc.
A multi-objective GA that identifies a few best individuals has been
developed at Ames [Lohn, et. al. 2000]. In this system, a population of
teachers evolve in parallel with the main population. Each teacher
consists of a desired level of performance for each objective. The fitness
of a teacher is inversely proportional to the number of individuals that
passes the teacher's test for all objectives -- or the worst possible fitness
if no individual can pass the teacher's test. The fitness of each individual
is proportional to the number of teacher-tests the individual passes. As
evolution progresses, the teacher tests become more difficult and the population
improves, but there is always one or a few individuals with the highest
fitness. These will be chosen for emigration.
Web-Centric Design
Components
We propose a master-slave architecture with immigrants passing from slave
to slave through the master. Communication is via a web server with a relational
database backend.
The design consists of seven elements: master, slave, web server, database
server, cycle scavenger, web browser, and charting applet. Figure 3 shows
their relationships:
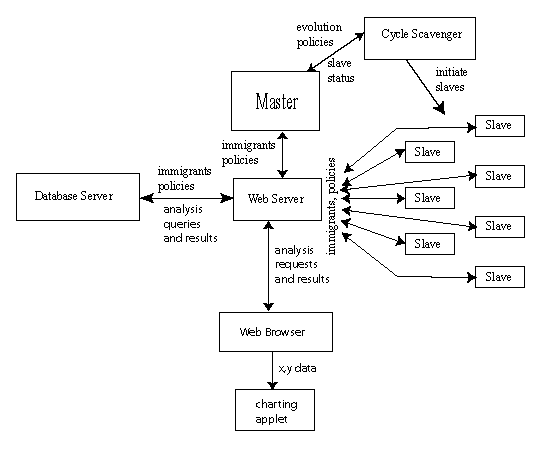
|
Figure 3: planned web-centric JavaGenes architecture.
|
-
Slaves: slaves manage a population. Slaves run a multi-objective GA until
killed. The GA uses [Lohn, et. al. 2000]'s technique where an evolving
population of teachers can be used to determine a few best individuals.
Periodically, slaves send the best individuals to the web server, check
for immigrants, and check for a new policy. Each of these may be at different
time intervals. If a new policy is provided, the current best individual
is sent to the database so improvement can be measured.
-
Master: manages a central database of individuals consisting of the current
pareto front and it's history. The master also determines where immigrants
go and the evolution policy of the slaves. The evolution/immigration policies
can evolve as the computation proceeds. A policy's fitness is the fitness
of the best individual the slave has produced and the improvement of the
best individual since the last policy change. The evolution policy includes:
-
Population size
-
Transmission operators, their parameters, and frequency of use
-
Method for choosing parents
-
Subset of objective to evolve
-
Immigrant and emigrant frequency
-
Frequency of policy change
-
etc.
-
Database server: off-the-shelf database server holding the pareto front
and it's history. The baseline design uses mySQL [http://www.mysql.com].
-
Web Server: off-the-shelf web sever. Except for initiation and termination,
all slave communication passes through the web server using SSL. PHP pages
implement the interaction. All access to the database must pass through
the web server. This architecture means that the slaves need no special
IO facilities on their hosts (other than internet access and standard Java
libraries). In particular, after initiation all IO is Cycle Scavenger independent.
-
Cycle Scavenger: off-the-shelf cycle scavenger (Condor, United Devices,
Entropia, Parabon, etc.). This element initiates, terminates and manages
the slave processes.
-
Web Browser: off-the-shelf web browser. This is used for requesting data
analysis (via SQL queries) and examining results.
-
Charting applet: modified off-the-shelf applet. This applet draws small
multiples (see figure 2) of two dimensional charts of the data returned
by SQL queries.
Performance
Large numbers of slaves have the potential to overwhelm the master, web
server, and/or database server with requests and information. For this
reason, every slave interaction with the web server will receive a time
in milliseconds as a response. This time is the minimum time before the
slave may re-contact the web server. By observing the load on the central
systems and changing this time value, the master can keep the load on the
central systems at reasonable levels. Also, rogue slaves may be detected
if they re-contact the web-server before the requested time.
Security
It will be possible for sophisticated individuals to feed bogus data to
JavaGenes. This must be detected and ignored. Also, it may be possible
to reverse engineer the JavaGenes slave code from the Java byte code sent
to the slaves. JavaGenes is well documented in the open literature and
may be distributed within the US, so while steps to prevent this problem
should be taken, absolute assurance is overkill. Finally, unauthorized
individuals may try to gather information on the computation by examining
IO. Since each slave sees only a tiny fraction of the computation,
and the results of the calculation are expected to be published in the
open literature, this problem also does not require iron-clad security.
There are a couple of possible attacks on proposed system:
-
An unauthorized party could take control of the code and/or data sent to
a slave. However, the Condor nodes are inside the NAS security walls and
United Devices encrypts all data sent to computational nodes. Therefor,
these data are only accessible in memory. Furthermore, Java byte code can
be obscured to make reverse engineering of the byte code difficult.
-
An unauthorized party could modify slaves to send incorrect data to the
master. The only possible truly wrong data is the fitness values for the
immigrants. However, the number of immigrants is tiny compared to the total
population under evolution (a ratio of perhaps 1/100,000 or greater). Therefor,
the master can simply re-evaluate the fitness (in this case independent
of the teachers). Also, immigrants can be sent via SSL to make reverse
engineering the format difficult.
-
An unauthorized party could flood the master with messages in a denal-of-service
attack. The timing approached mentioned above can limit this, although
any web server is subject to such an attack.
-
An unauthorized party could attempt to corrupt the database directly, without
going through the web server. Currently, the NAS mySQL server only allows
access from build-dev.nas.nasa.gov. Furthermore, mySQL database privileges
may be set to minimize the capabilities of each part of the system. Namely:
-
Slaves may only add and retrieve from/to the immigrant tables and retrieve
from policy tables
-
Master may only retrieve/delete from immigrant tables, add to history tables,
add/delete/retrieve from policy tables, create databases and tables.
-
Analysis may only retrieve data.
Security is also an issue for the proposed analysis capabilities. While
unauthorized reading of the data is of relatively little concern, since
it will be published in the open literature eventually anyway, modification
of the database and denial of service attacks could be problematic. These
can be addressed by password protecting the relevant web pages and by limiting
access to the machines used by the author and his collaborators.
IPG Design
It is possible to implement the design as a message passing system by replacing
the web-server with a message passer for the compute portion of the system.
The IPG currently provides an implementation of MPI 1.1 [http://www-unix.mcs.anl.gov/mpi]
called MPICH-G2 [http://www.niu.edu/mpi]. However, MPI 1 does not
allow processes to be added or removed once an application has started,
so MPICH-G2 cannot be used in a cycle-scavenging environment. MPI 2 does
allow for dynamic changes to an application's processor so it may work.
Although MPICH-G2 implements part of MPI 2, it does not implement a dynamic
process pool.
Also, the MPI standard is explicitly intended for C and FORTRAN, not
Java, programs, so a JNI (Java Native Interface) is necessary for Java
programs to connect to an MPI library. This makes applications somewhat
less platform independent since a separate native code library is necessary
for each architecture. Nonetheless, if a MPI 2 implementation were
available as part of cycle-scavenger compute node implementation one could
replace the web server with a central message handler. However, as
Java has excellent communication facilities and MPI has a very small market
share, it may be difficult to convince commercial vendors to add MPI 2
to their cycle-scavenging communication facilities.
Another approach to using current IPG capabilities would be to use globus-io
[http://www.globus.org/v1.1/io/globus_io.html] to pass data to and from
a central machine. A polling mechanism could then insert the data
into the database using the web server and/or a more direct interface.
globus_io provides both security and non-blocking IO. Again, as with
MPI, globus-io is intended for C and FORTRAN programers so a JNI interface
would be necessary. Globus-io could be added to the Condor installation
and be reliably available to Condor programs, however it may be difficult
to convince commercial companies to implement globus-io as they already
have their own mechanisms for compute nodes to communicate with their central
servers.
In the next few months, it seems impractical to use globus communication
libraries for cycle-scavenging applications. However, the cycle-scavenging
community seems to be ripe for some kind of standardization, and perhaps
the IPG can help. Condor is already moving towards IPG integration
with Condor-g; and NAS is planning to make the Condor pool accessible
to globus jobs on an experimental basis (there are some security concerns).
Entropia has announced they will integrate their product with the IPG [http://www.entropia.com/release_09272001.asp].
As NASA works with the vendors there should be opportunities to move towards
a more integrated set of systems that may even allow applications to take
advantage of multiple cycle-scavengers. For the present effort, certain
tasks (e.g., immigrant validation) could be performed on trusted Condor
nodes while the bulk of evolution continues on untrusted United Devices
nodes. The lessons learned should be valuable in developing interoperability
standards for cycle-scavengers in the same manner that the IPG is developing
interoperability for more conventional supercomputers.
There is one area the IPG could help with in the near future.
If both Condor and United Devices were accessible to globus jobs, then
the master would see a uniform environment for initiating and terminating
jobs. NAS Condor pool accessibility to globus jobs will be tested
in the next few months, and it is possible, in principle, to build a globus
front end for the United Devices MetaProcessor. Although the details of
Entropia/globus integration are not yet known, they almost certainly include
running globus jobs on Entropia system. Having all systems implement globus
job control would simplify job initiation and management code in the master.
Conclusion
In the next several months JavaGenes is expected to evolve from a simple
genetic algorithm with disc IO run on the NAS Condor pool to a web-centric
application distributed across the Internet with a relational database
for storage using IPG process management. This change is driven by the
need to move from execution on a pool of a few hundred UNIX workstations
in one location to execution on up to a hundred thousand PCs scattered
around the world. This will allow real world experience with massively
distributed cycle-scavenging applications to drive IPG standardization
in this area.
Acknowledgments
I would like to thank the NAS IPG Project [http://www.ipg.nasa.gov/] for
funding this work. Thanks to Eric Langhirt for reviewing the manuscript.
References
[Akira
and Meng-Sing 2001] "Multiobjective Optimization of Rocket Engine Pumps
Using Evolutionary Algorithm," Oyama Akira and Liou Meng-Sing
, NASA/TM-2001-211082 or AIAA2001-2581, CD-ROM Proceedings of 15th AIAA
Computational Fluid Dynamics Conference, Anaheim, CA, 11-14, June, 2001.
[Globus,
et. al 1999] "Automatic molecular design using evolutionary techniques,"
Al Globus, John Lawton, and Todd Wipke, Nanotechnology, Volume 10,
Number 3, September 1999, pp. 290-299.
[Globus,
et. al 2000a] "JavaGenes: Evolving Graphs with Crossover," Al Globus,
Sean Atsatt, John Lawton, and Todd Wipke, 2000.
[Globus,
et. al 2000b] "JavaGenes and Condor: Cycle-Scavenging Genetic Algorithms,"
Al Globus, Eric Langhirt, Miron Livny, Ravishankar Ramamurthy, Marvin Solomon,
and Steve Traugott, Java Grande 2000, sponsored by ACM SIGPLAN, San Francisco,
California, 3-4 June 2000.
[Globus,
et. al 2001] "JavaGenes: Evolving Molecular Force Field Parameters,"
Al Globus, Charles Bauschlicher, Sandra Johan, and Deepak Srivastava, abstract
accepted for oral presentation at the Ninth Foresight Conference on Molecular
Nanotechnology, 9-11 November 2001 in Santa Clara, California.
[Goux, et. al 2000]
"An Enabling Framework for Master-Worker Applications on the Computational
Grid," J. P. Goux, S. Kulkarni, J. T. Linderoth, and M. E. Yoder,
Proceedings of the Ninth IEEE International Symposium on High Performance
Distributed Computing, 2000.
[Holst and Pulliam 2001] Terry L. Holst and Thomas H. Pulliam,
"Aerodynamic Shape Optimization Using A Real-Number-Encoded Genetic
Algorithm" AIAA 2001-2473, June, 2001
[Laughlin
and Chambers 2001] "Short Term Dynamical Interactions among Extrasolar
Planets," Laughlin and Chambers, Astrophysical Journal Letters v. 551,
p. L109., 11 April 2001.
[Litzkow, et al. 1988] M. Litzkow, M. Livny, and M. W. Mutka, "Condor
- a Hunter of Idle Workstations,'' Proceedings of the 8th International
Conference of Distributed Computing Systems, pages 104-111, June 1988.
See http://www.cs.wisc.edu/condor/.
[Lohn and Colombano 1998] "Automated Analog Circuit Synthesis Using
a Linear Representation," Jason D. Lohn and Silvano P. Colombano, Second
International Conference on Evolvable Systems: From Biology to Hardware,
Springer-Verlag, 23-25 September 1998.
[Lohn, et. al. 2000] "A Comparison of Dynamic Fitness Schedules for
Evolutionary Design of Amplifiers," Jason Lohn, Gary Haith, Silvano Colombano,
and Dimitris Stassinopoulos, Proceedings of the First NASA/DoD Workshop
on Evolvable Hardware, Pasadena, CA, IEEE Computer Society Press, 1999,
pages 87-92.



