Figure 1: Crossover
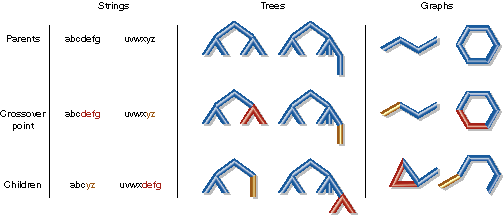
Genetic algorithms differ in their representation of solutions. Bit string representations were used in the first genetic algorithms [Holland 1975], but arrays of floating point numbers, special symbols that generate circuits [Lohn and Colombano 1998], robot commands [Xiao, et al. 1997], and many other symbols may be found in the literature. Strings may be of fixed or variable length. Trees can also be evolved [Koza 1992]. This is usually called genetic programming, because trees are particularly useful for representing computer programs. Many molecules contain cycles, which chemists call rings, and strings and trees don't contain cycles. Therefore, we took the unusual approach of evolving graphs. Graphs are a set of vertices (for example, atoms) and a set of edges (for example, bonds), each of which connects two vertices. In this paper, the term graph does not refer to a two dimensional image used for data presentation. Figure 1 depicts crossover using strings, trees and graphs.
Figure 1: Crossover

| Crossover can be applied to strings, trees, and graphs. Note that only graph crossover requires multiple break points and that recombination must work on fragments with different numbers of broken edges. Particularly with large multi-ring molecules, these complex data structure manipulations are more error prone in C/C++/FORTRAN than in Java. |
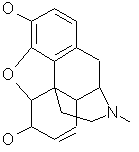
Evolving graphs involves randomly splitting arbitrary graphs into two fragments and then combining fragments. Splitting a complex graph, such as morphine (figure 2) involves data structure manipulations that can easily result in various bugs. Java was chosen as the implementation language to minimize these problems.
Figure 2: Morphine
JavaGenes has been used for pharmaceutical drug and digital circuit design. One approach to drug design is to find molecules similar to good drugs. Ideally, a candidate replacement drug is sufficiently similar to have the same beneficial effect, but is different enough to avoid negative side effects. To use JavaGenes for similarity-based drug discovery we need a good similarity measure that can score any molecule. [Carhart, et al. 1985] defined such a similarity measure, all-atom-pairs-shortest-path, and searched a large database for molecules similar to diazepam. We use a closely related similarity technique to evolve a population of molecules towards a target drug molecule.
For an excellent review of genetic algorithms and related techniques as of Spring 1997, see [Baeck 1997].
Embarrassingly parallel programs are a natural match for Condor [Litzkow, et al. 1988]. Condor is a software system that creates a High Throughput Computing [Livny and Raman 1999] environment by effectively harnessing the power of a cluster of UNIX workstations on a network. Although Condor can manage a dedicated cluster of workstations, a key appeal of Condor is its ability to effectively harness non-dedicated, preexisting resources in a distributed ownership setting such as machines sitting on people's desks in offices and labs. We ran JavaGenes on the NAS Condor pool. NAS is the primary NASA supercomputer center [NAS]. Approximately 200 workstations, purchased and used for software development, visualization, email, document preparation, etc., are available for batch processing during idle times. The Condor daemons watch these 200 workstations. When a workstation has been idle for 2 hours, a job from the batch queue is assigned to the workstation and will run until the workstation detects a keystroke, mouse motion, or relatively high non-Condor CPU usage. At that point, the job will be removed from the workstation and placed back on the batch queue. As mentioned before, it's not uncommon to have a few hundred JavaGenes jobs in the queue.
Because a JavaGenes job running under Condor may be vacated (removed from the workstation) at any time, the job must save state (checkpoint) periodically. Condor provides a generic checkpoint/restart facility, but for reasons discussed below, we could not use this facility for JavaGenes. Checkpointing was initially implemented using standard Java serialization. Conceptually, this is relatively simple since the state of a genetic algorithm is simply the current population. However, standard Java serialization turned out to be a serious, but fixable, performance problem, as others have discovered [Wims and Xu 1999]. See the section below on serialization performance.
Remote system calls provide a uniform environment to a job running on any workstation on a network. The Condor runtime library replaces system calls with remote procedure calls to a shadow process running on the workstation that submitted the job. The shadow makes the system call on behalf of the remote process and returns the results. For example, the open system call sends the name of the desired file to the shadow, which searches for it on the submitter's home workstation. Subsequent read and write calls access that file over the network. The result is that the job sees the same file-system environment regardless of where it runs, and all file output is captured in files on the submitting workstation.
Checkpointing is also implemented by the Condor runtime library. Each Condor job is run under the control of a starter process. When a workstation needs to be appropriated for another purpose, the starter sends a terminate signal to the application process. The Condor library catches this signal and sends a complete dump of the state of the process back to the submitting machine, where it awaits its turn to be restarted on another worker machine. This checkpoint file includes a binary dump of the entire virtual memory image of the process. It also includes a record (collected by the remote system calls) of the current state of the process' interaction with the operating system kernel. For example, for each file opened by the job, the checkpoint file records the name of the file and the current offset within the file (the "seek pointer"). Condor can also be instructed to send a "checkpoint" signal to the starter at periodic intervals. The starter responds to this signal by suspending the application, checkpointing it, and then allowing it to continue. Under some circumstances, Condor may send other signals to the starter, asking it to suspend the application, resume it, or kill it without giving it a chance to checkpoint.
Neither remote system calls nor checkpointing require any source-level modifications to the application program, but they do require it to be re-linked with a special version of the system libraries. They also impose some restrictions on the set of system services available to the job. In particular, Condor does not currently support Standard jobs that use kernel-level threads. Programs that cannot be re-linked or that do not meet these requirements must be run in the Vanilla universe. An arbitrary executable program can be run as a Vanilla job, but any I/O operations will access the file system of whatever machine the job happens to be running on, and if the workstation is pre-empted (for example, by an interactive user), the job is simply killed and restarted from the beginning on another workstation. At NAS, in general, Condor jobs do not have permission to use the local disk on the worker workstation.
Most of the JavaGenes runs described in this paper used the Vanilla universe. The Java Virtual Machine (JVM) from SGI and Sun was available to us only in binary (pre-linked) form. We tried the Kaffe open-source JVM, but found that it had bugs that prevented us from running JavaGenes correctly for more than a few generations. Moreover, both JVM's use certain facilities -- notably kernel-level threads -- that are not supported in Condor Standard jobs. Fortunately, the remote system call facility was not necessary in our environment, since we use the Network File System (NFS), which provides a uniform interface to files from all workstations. In this environment, the "job" submitted to Condor is a tcl script that calls the JVM, with the name of the application class supplied as a command-line argument. From the point of view of the JVM, the class files that constitute the application are simply data files that appear to be on the local disk of the worker machine through the magic of NFS. Similarly, NFS is used to create output files on the submitting workstation.
The lack of automatic checkpointing was a more serious problem. As mentioned earlier, we tried two different application-specific checkpointing strategies. Both involved periodically invoking a checkpoint method that saves the state of the computation into a file. JavaGenes invokes this method after each new child is created. If a workstation is preempted before the program finishes, it is killed and restarted "from the beginning." However, at startup, JavaGenes jobs look for a checkpoint file (via NFS) so they can initialize state and continue from the last checkpoint. Thus, a job that is killed and restarted loses only the work it did between its most recent checkpoint and the time it was killed.
To provide better support for JavaGenes and other Java Grande applications,
the Condor project has been developing a Java universe. To run under this
universe, a Java program must implement the Checkpointable interface:
public interface Checkpointable extends Serializable
{
void start(String[]
arguments);
void restart();
void beforeCheckpoint();
void afterCheckpoint();
void setCheckpointer(Checkpointer
c);
}
Each universe has its own kind of starter. The starter for the Java universe is written in Java and extends java.lang.ClassLoader. A "job" in the Java universe is a class file. We assume that each worker machine has a JVM installed locally, but otherwise do not require any network file system or uniform file environment. The Java starter loads all required classes over the network by communicating with a Java version of the shadow using the Java RMI (Remote Method Invocation) interface. The Java starter creates an instance prog of the application program class and calls either prog.start() or prog.restart(), depending on whether there is an existing checkpoint file from an earlier run of this job. The starter also creates an instance cp of a Checkpointer object, passing prog to its constructor, and calling prog.setCheckpointer(cp) so that prog and cp can refer to each other. Prog can also access cp through static methods in Checkpointer as long as there is only one instance of Checkpointer. When the starter receives a terminate or checkpoint signal, it calls cp.checkpointWhenPossible(), which sets a flag indicating that a checkpoint has been requested. It does not force an immediate checkpoint because the application object may not be in a "quiescent" state in which checkpointing is convenient. The application itself is expected to call cp.ok() periodically. If no checkpoint has been requested, this method simply returns without doing anything. However, if a checkpoint request is pending, the checkpointer uses Java serialization to save the state of the object by calling writeObject(prog). It also calls prog.beforeCheckpoint() before the checkpoint and prog.afterCheckpoint() after.
The writeObject method saves all of the non-transient fields of the object, as well as all objects pointed to by those fields, all fields of those objects, etc. It does not, however, save anything from the runtime stack -- that is, the values of local variables. It is the responsibility of the application to update the non-transient fields of the application object to reflect the complete state of the application either before calling cp.ok or in the method beforeCheckpoint. If these updates are costly, they should be in beforeCheckpoint because this method is only called if the checkpoint is actually performed. The beforeCheckpoint and afterCheckpoint methods also provide hooks for application-specific performance monitoring, such as determining the amount of time spent checkpointing. A typical application might look like this:
class Application implements Checkpointable {
private Checkpointer
checkpoint;
private GlobalState
state;
private int lastStepCompleted;
private transient Vector
intermediateResults;
private int iterations;
// set by initializeState
private PerformanceStatistics
statistics;
public void setCheckpointer(Checkpointer
checkpoint) {
this.checkpoint = checkpoint;
}
public void start(String[]
args) {
state.initialize(args);
intermediateResults = new Vector();
iterateFrom(0);
}
public void restart()
{
intermediateResults = new Vector();
state.restore(intermediateResults);
iterateFrom(lastStepCompleted);
}
private void iterateFrom(int
start) {
for (int i = start; i < iterations; i++) {
intermediateResults.add(oneStepOfAlgorithm());
lastStepCompleted = i;
checkpoint.ok();
}
printResults();
}
public void beforeCheckpoint()
{
statistics.startTimer();
state.update(intermediateResults):
}
public void afterCheckpoint()
{
statistics.stopTimer();
}
// Methods initializeState,
oneStepOfAlgorithm,
// startTimer, etc.
omitted for brevity.
}
The Java universe provides several advantages over the Vanilla universe for Java applications:
One other fairly serious problem arose. In the initial implementation, JavaGenes started a new random number generator after restart. Because of this, evolution did not follow the same path taken after the last checkpoint. In other words, the genetic algorithm was controlled by different random numbers and therefore searched a different part of the search space. That made the algorithm appear more efficient than it was, because the number of generations to find a specific target was used as the efficiency measure. Consider the case where a job ran for 100 generations up to a checkpoint and 20 more generations before being killed. When the job was restarted, 20 different generations would be constructed and one of the new generations, say the 10th, might find the target. Therefore, it would appear as if the target was found in 110 generations, but actually 130 were tried. Thus, jobs might repeatedly search the region around the last checkpoint and stop if a solution were found, reporting an inaccurately small number of generations to completion. It was therefore necessary, at job restart time, to restart the random number generator with the same seed and then execute the random number generator the number of times it was executed up to the last checkpoint. This should have insured that a job would follow the same evolutionary path regardless of checkpoint history. Unfortunately, although this fix worked properly when a job was restarted on the same machine, different results were observed in normal execution on the Condor pool, where jobs frequently move between machines. These changing evolutionary paths were presumably caused by differences in the Java libraries on different versions of the SGI operating system. This may change the order of certain lists in JavaGenes and cause the same random number to pick different items from these lists. Fortunately, only about 6% of the generations were repeated once checkpointing performance was improved, suggesting that the effect was fairly minor. Because checkpointing was fast, computations would usually proceed to the next checkpoint before being restarted.
In short, we were happy with Java for this application. While there is some run-time penalty, more rapid code development and more reliable end-product software is well worth the extra CPU cycles. Since cycles are always getting cheaper, and programmers and support staff seem to be getting more expensive, we expect Java to do well in the coming years.
[Carhart, et al. 1985] Raymond Carhart, Dennis H. Smith, and R. Venkataraghavan, "Atom Pairs as Molecular Features in Structure-Activity Studies: Definition and Application," Journal of Chemical Information and Computer Science, volume 23, pages 64-73.
[Globus, et al. 1999] Al Globus, John Lawton, and Todd Wipke, "Automatic Molecular Design Using Evolutionary Techniques," Sixth Foresight Conference on Molecular Nanotechnology, Sunnyvale, California, November 1998 and Nanotechnology, volume 10, number 3, September 1999, pages 290-299.
[Globus, et al. 2000] Al Globus, Sean Atsatt, John Lawton, and Todd Wipke, "JavaGenes: Evolving Graphs with Crossover," in preparation.
[Holland 1975] John H. Holland, Adaptation in Natural and Artificial Systems, University of Michigan Press.
[Koza 1992] John R. Koza, Genetic Programming: on the Programming of Computers by Means of Natural Selection, MIT Press, Massachusetts.
[Litzkow, et al. 1988] M. Litzkow, M. Livny, and M. W. Mutka, "Condor - a Hunter of Idle Workstations,'' Proceedings of the 8th International Conference of Distributed Computing Systems, pages 104-111, June1988. See http://www.cs.wisc.edu/condor/.
[Livny and Raman 1999] M. Livny and R. Raman, "High Throughput Resource Management," Computational Grids: The Future of High-Performance Distributed Computing, edited by Ian Foster and Carl Kesselman, published by Morgan Kaufmann, 1999.
[Lohn and Colombano 1998] Jason D. Lohn and Silvano P. Colombano, "Automated Analog Circuit Synthesis Using a Linear Representation,'' Second International Conference on Evolvable Systems: From Biology to Hardware, Springer-Verlag, 23-25 September 1998.
[NAS] http://www.nas.nasa.gov/home.html.
[Tunkelang 1998] Daniel Tunkelang, A Numerical Optimization Approach to Graph Drawing, Dissertation, Carnegie Mellon University, School of Computer Science, December 1998.
[Wims and Xu 1999] Brian Wims and Cheng-Zhong Xu, "Traveler: A Mobile Agent Infrastructure for Wide Area Parallel Computing," ACM 1999 Java Grande Conference, San Francisco, California, 12-14 June 1999.
[Xiao, et al. 1997] Jiang Xiao, Zbigniew Michalewicz, Lixin Zhang, and
Krzysztof Trojanowski, "Adaptive Evolutionary Planner/Navigator for Mobile
Robots," IEEE Transactions on Evolutionary Computation, volume 1,
number 1, pages 18-28, April 1997.