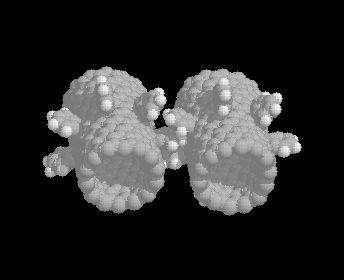
A typical gear configuration.
Al Globus, MRJ, Inc. at NASA Ames Research Center and Richard Jaffe, NASA Ames Research Center.
The first systems we have investigated are various gears built out of single walled carbon nanotubes with o-benzyne groups attached to form the teeth. [Thess 96] has demonstrated a 70% yield in carbon nanotube production so the tube should be synthetically accessible, although [Thess 96] generated (10,10) tubes whereas most of our simulations use (14,0) tubes. [Hoke 92] has shown that benzyne reacts with C60 to form a 1-2 bond between six membered rings and quantum calculations [Jaffe 96a] suggest that a similar reaction should take place on carbon nanotubes, although 1-4 bonds are slightly preferred. Adding aromatic rings to the tube should give us relatively stiff molecular gear teeth, and this has proved to be the case [Han 96].

A typical gear configuration.
Using the NanoDesign design and simulation software described below,
[Han 96] has shown that --
assuming you believe the force field -- a number of gear
and gear/shaft systems will function mechanically in a vacuum. These
simulations used a software thermostat and motor, but there is reason
to believe that physical implications of these
functions can be provided. Preliminary simulations suggest that cooling is possible using an
inert atmosphere. Experimental evidence (Sunny Bains reports in
Science , volume 273, 5 July 1996, p. 36 on upcoming papers) and
simulation [Tuzun 95] suggest that lasers may be used to rotate the
gears. The tube is functionalizing with positive and negative charges
in appropriate locations and the lasers are used to create a rotating
electric field.
The current system consists of a parallelized FORTRAN program to simulate
hydrocarbon systems. Supramolecular conformations come from xyz files
(the force field does not require a bond network in the input) produced
by a c++ and tcl program using the tcl_c++ interface generator.
The software also creates FORTRAN files with indices into an array of atoms
indicating where each component (e.g., gear teeth) begins and ends. The
user creates tcl files with tcl functions to create and modify c++ objects.
For example, this tcl fragment creates a buckytube:
Design Software
The simple molecular machines simulated so far can be easily designed
and modeled using ad hoc software and molecule development. However,
to design complex systems such as the molecular assembler/replicators
envisioned by the
NASA Ames Computational Molecular Nanotechnology Project
[Globus 96b], a more sophisticated
software architecture will be needed. The current NanoDesign software
architecture is a set of c++ classes with a tcl front end for interactive
molecular gear design. Simulation is via a parallelized FORTRAN program
which reads files produced by the design system.
We envision a future architecture centered around
an object oriented database of molecular machine components and systems
with distributed access via CORBA from a user interface based on a
WWW universal client.
Current Software Architecture
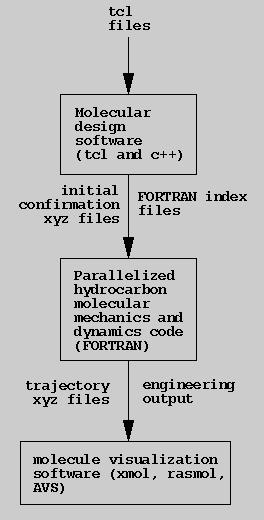
Current NanoDesign software architecture.
# create a buckytube
set tube [aBuckytube]
# it will be 14,0 tube
$tube setRingCircumference 14
# make it 21 rings long
$tube setRingLength 21
# set the FORTRAN variable name for the tube
$tube setVariableName "tube"
# tell c++ to create the tube
$tube build
# write the confirmation into a file
$tube writeXyz "tube.xyz"
# write the FORTRAN declarations and index assignments into a file
$tube writeFORTRANVariables "tube.f"
See here for details on the FORTRAN output.
The
Visualization Toolkit project
[Schroeder 96] discovered that a tcl interface to a
large c++ class library can require substantial programmer effort to
write the glue that allows tcl to control c++ classes. The
vtk project avoided this by writing a partial c++ header file parser
that reads the c++ header file for a class and automatically generates the
tcl interface code. We wanted more control over which c++ member functions
were tcl accessible, so the tcl_c++ system requires a file for each c++
class to define which member functions, variables, and constants are tcl
accessible. This file is read by a tcl interpreter with tcl procedures
defined to generate c++ code to allow another tcl interpreter to control
the c++ class in question. Fortunately, although tcl_c++ itself was
hard to program, it is easy and convenient for a programmer to use.
For details of tcl_c++ see
here.
The current NanoDesign molecular design software appears to the
user as an interpreted language based on tcl. This is very
effective for design of simple parts and systems. To design
and computationally test complex replicators will require a more
sophisticated system similar to the mechanical CAD systems available
in the commercial marketplace. Furthermore, it would be of substantial
practical advantage if the design team could be geographically dispersed.
Therefore, we are investigating an software architecture based on a
universal client (for example, a WWW browser), CORBA distributed objects,
an object oriented database, and encapsulated computational chemistry
legacy software. We are also interested in using command language
fragments to control remote objects. Software that communicates
this way is sometimes called agents [Genesereth 94].
Recently, Netscape, Inc. announced that the netscape
WWW browser would be made CORBA (see below) compliant offering a standard
way to communicate between application code loaded by the browser and
databases and computational chemistry software resident on servers and
supercomputers. Previously, only the stateless http protocol was
available to web browsers. Hopefully, other companies in the extremely competitive
WWW browser market will follow suit.
These developments suggest that a single program can function as the
user interface for a wide variety of applications, including computational
nanotechnology. These applications
load software (e.g. Java applets and JavaScript) into the browser when
the user requests it. The applications then communicate with databases
and remote objects (such as encapsulated legacy software) to meet user
needs.
To companion papers.
Proposed Future Software Architecture
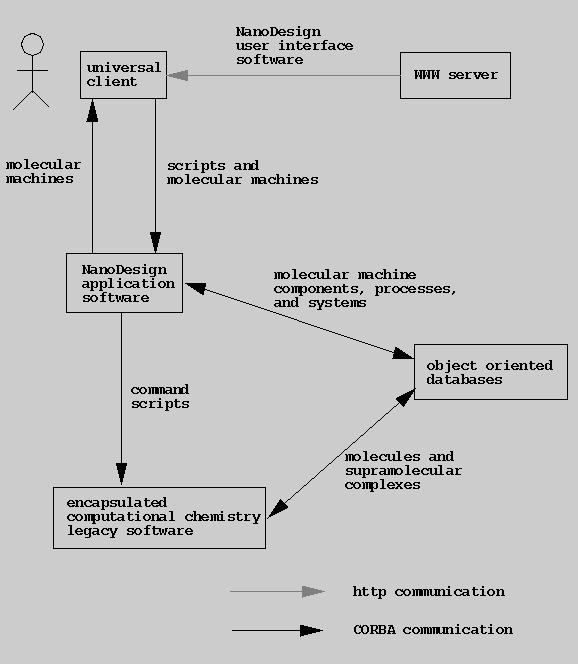
Future distributed NanoDesign software architecture.
Note that each box may represent many instances distributed onto almost any machine.
Universal Client
With the advent of modern WWW browsers implementing languages such as
Java and JavaScript, it is possible to write applications using these
browsers as the user interface. This saves development time since most
user interface functionality comes free, integration with the WWW is trivial,
and the better browsers run on a wide variety of platforms so portability
is almost free. VRML can be used for 3D graphics and plug-ins such as the
recently announced Biosym/MSI, Inc. molecule browser provide crucial
functionality without much work.
CORBA (Common Object Request Broker Architecture)
The universal browser is of little use in developing complex molecular
machines if it cannot communicate with databases of components
and systems and invoke high performance codes on fast machines to do
the analysis. CORBA, a distributed object standard developed by the
OMG (Object Management Group), provides a means for distributed objects --
for example the universal browser application, a database containing
an evolving molecular machine design, and simulation codes -- to communicate
and control each other. The simplest description of CORBA is that each
object is represented by an interface described by the CORBA IDL
(interface description language). Operations and data defined in the
IDL may be accessed by other CORBA objects on the network. System software
(called ORBs -- object request brokers) is responsible for communicating
between objects whether they be on the same machine or widely distributed.
See [Siegel 96] for a description of CORBA.
Object Oriented Database
To develop complex molecular machines, databases of components and processes
as well as complex databases describing individual systems will be required.
Object oriented databases appear to be better than relational databases
for design systems for products
such as aircraft and molecular machines.
Encapsulated Computational Chemistry Legacy Software
Like most research centers, NASA Ames has a number of very
capable codes that do not fit the object model.
However, it is often possible to create a c++ object that
'encapsulates' the legacy software. That is, the c++ object
has methods that reformat their parameters, execute the
legacy software, reformat the result and return it.
When the legacy software does IO, the encapsulating object
must intervene between the legacy software and the CORBA system.
This technique allows existing codes to operate within an object
oriented framework with minimal modification.
Agent Style Communication
In this context, agent software means software components that
communicate by sending programs to each other. When each component
is controlled by a command language, this is relatively easy to
implement. Thus, a user interface component could control the
tcl/c++ design software by writing a tcl command file and sending
it to the design software for execution. This approach to software
is powerful but not yet well understood.
Conclusions
The NanoDesign software is intended to design and test fullerene based
hypothetical molecular machines and components. The system is in an
early stage of development. Presently, tcl provides an interpreted
interface, c++ objects represent design components, and a parallelized
FORTRAN program simulates the machine. In the future, an architecture
based on distributed objects is envisioned. A key requirement for this
vision is a standard set of interfaces to various computational chemistry
capabilities (e.g., force fields, integrators, etc.). A standard
set of interfaces would allow vendors to supply small, high quality
components to a distributed system. If you're interested in helping
establish these standards, please contact the author at globus@nas.nasa.gov.
Acknowledgments
I would like to thank my colleagues at the
Molecular Engineering Laboratory at the
University of California at Santa Cruz, led
by Dr. Todd Wipke, for many fruitful discussions
and an environment tremendously conducive to
molecular design.
 Web Work: Al Globus
Web Work: Al Globus